Introduction
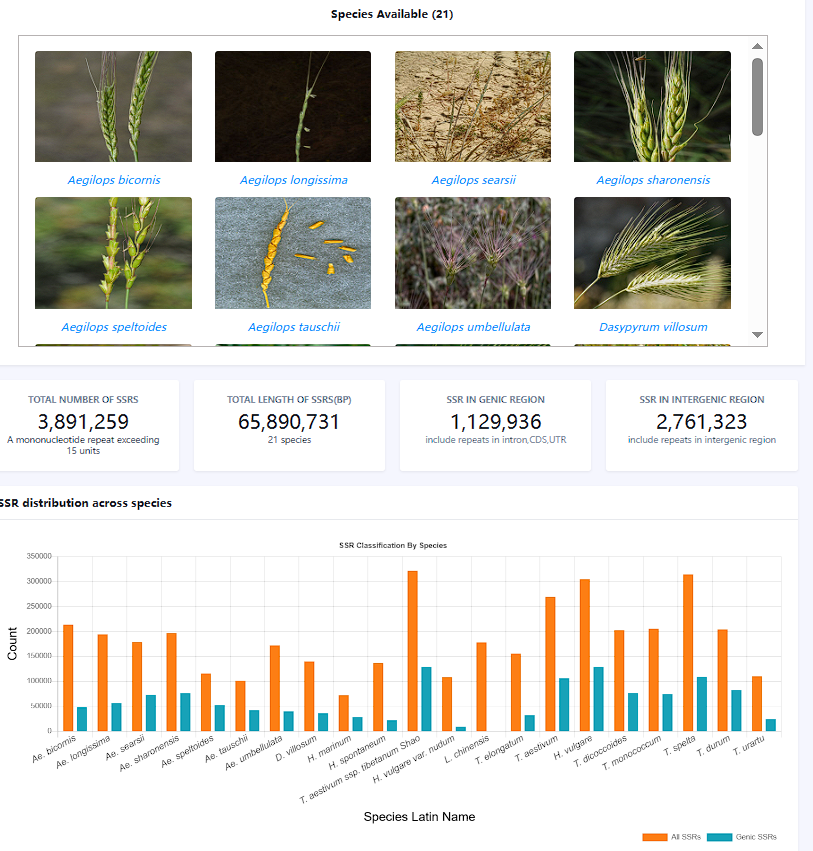
A total of 3,891,259 microsatellites were predicted from 21 Triticeae species for the development of the TriticeaeSSRdb web-resource.
The web server contains six tabs. Home, Species, Tools, Help, and Contact.
- On the home page, you can view the distribution of the number and type of microsatellites
were predicted from 21 species in a chart
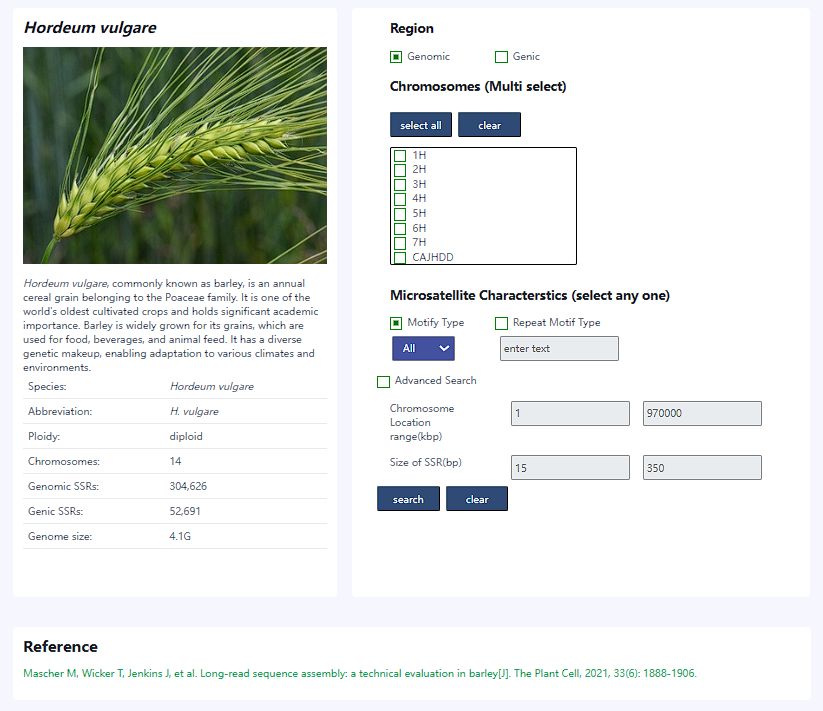
- The Species page provides information about the selected species on left and search options
on the right side. In silico predicted markers can be searched by selecting genic or
genomic, chromosome, along with motif type, repeat type, length, and location in the genome.
- The Tool page provides two tabs—prediction and blast.You can input sequences to predict
microsatellites and design primers and use local blast to align user query sequences with
the genome
Search Microsatellites and design primers
Select the species you want to search for SSR, you can browse introduction and distribution of SSR types for selected species.
- searched by selecting genic or genomic
- searched by different chromosome
- searched by motif type, repeat type, length, and location in the genome
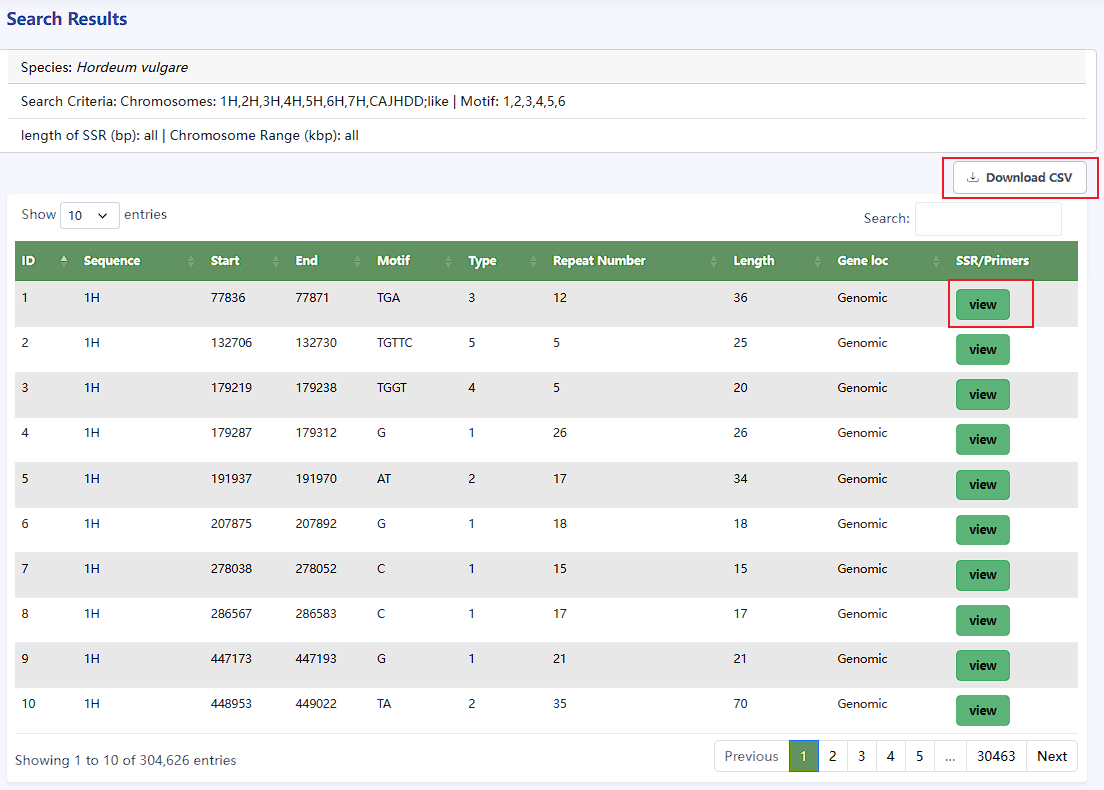
Based on the search query, the SSR and information will be displayed on the result page.
- You can click the Download CSV button to download the microsatellite locus information file of the selected species
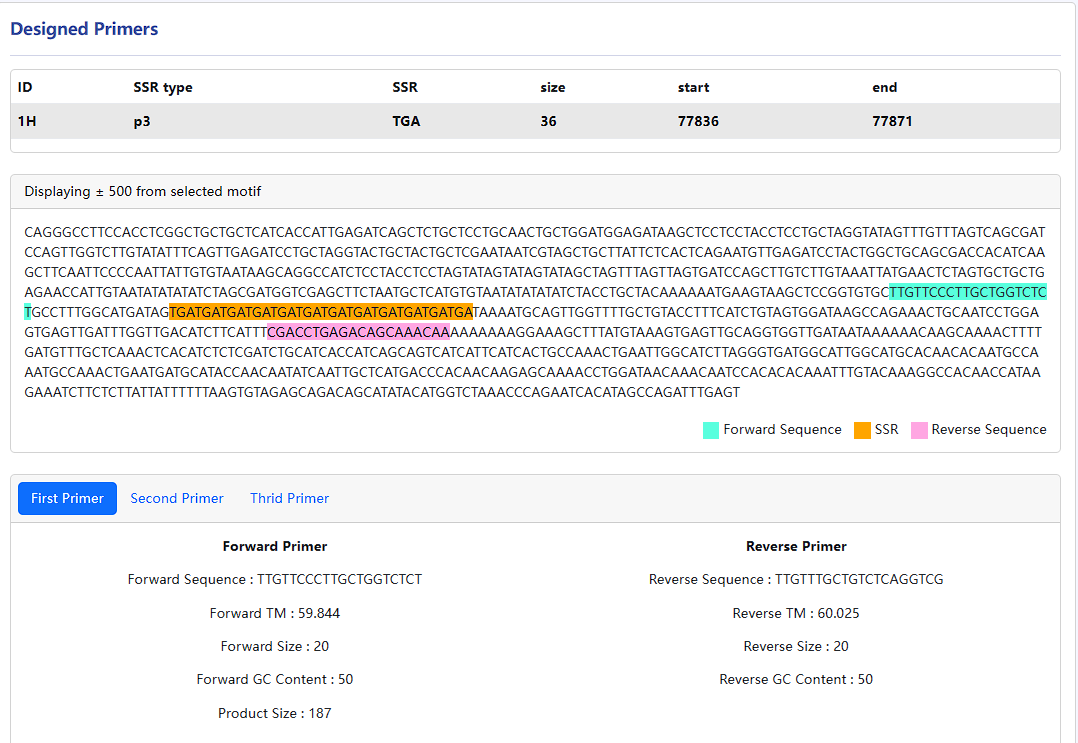
- Click the view button in a row to design primers for the flanking sequences of the microsatellite.
The repeat and designed primer are displayed in the sequence extracted with 500 bp upstream and downstream of the repeat.
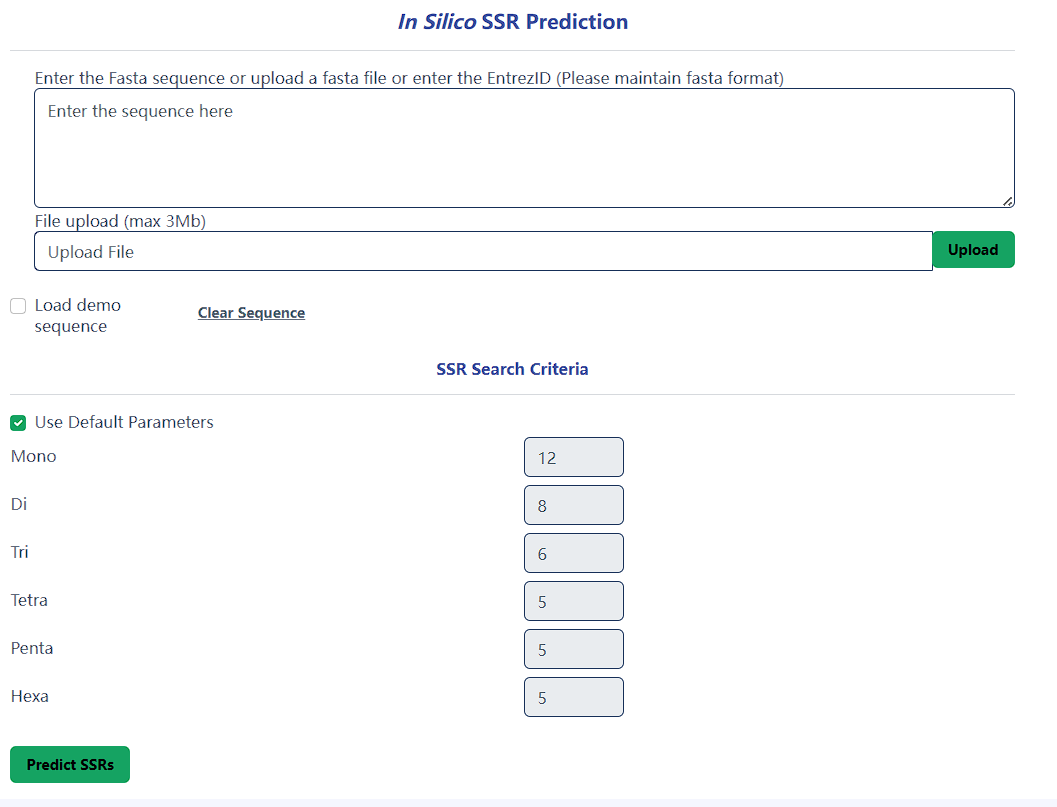
SSR predictor tool
You can input sequences to predict microsatellites and design primers
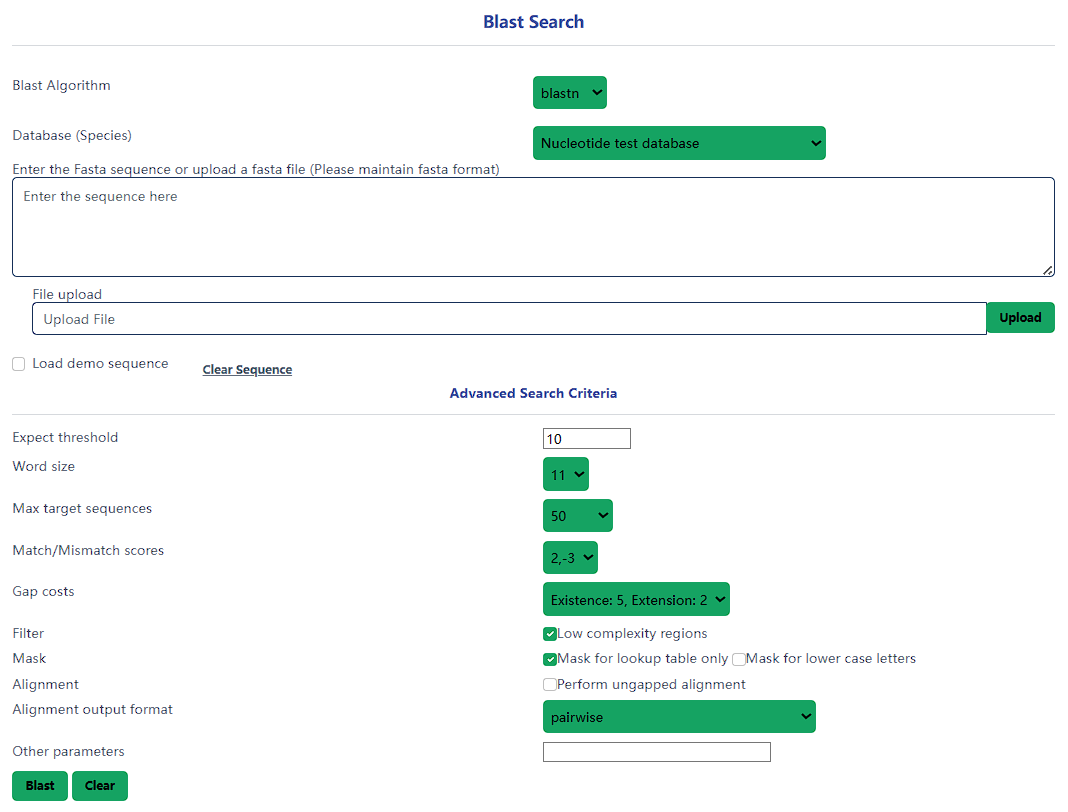
Local NCBI blast
You can use local blast to align user query sequences with the genome